The dscore() function estimates the following quantities: D-score,
a numeric score that quantifies child development by one number,
Development-for-Age Z-score (DAZ) that corrects the D-score for age,
standard error of measurement (SEM) of the D-score.
dscore(
data,
items = names(data),
key = NULL,
population = NULL,
xname = "age",
xunit = c("decimal", "days", "months"),
prepend = NULL,
itembank = NULL,
metric = c("dscore", "logit"),
prior_mean = NULL,
prior_mean_NA = NULL,
prior_sd = NULL,
prior_sd_NA = NULL,
transform = NULL,
qp = NULL,
dec = c(2L, 3L),
relevance = c(-Inf, Inf),
algorithm = c("current", "1.8.7"),
verbose = FALSE
)
dscore_posterior(
data,
items = names(data),
key = NULL,
population = NULL,
xname = "age",
xunit = c("decimal", "days", "months"),
prepend = NULL,
itembank = NULL,
metric = c("dscore", "logit"),
prior_mean = NULL,
prior_mean_NA = NULL,
prior_sd = NULL,
prior_sd_NA = NULL,
transform = NULL,
qp = NULL,
dec = c(2L, 3L),
relevance = c(-Inf, Inf),
algorithm = c("current", "1.8.7"),
verbose = FALSE
)Arguments
- data
A
data.frameormatrixwith the data. A row collects all observations made on a child on a set of milestones administered at a given age. The function calculates a D-score for each row. Different rows can correspond to different children or ages.- items
A character vector containing names of items to be included into the D-score calculation. Milestone scores are coded numerically as
1(pass) and0(fail). By default, D-score calculation is done on all items found in the data that have a difficulty parameter under the specifiedkey.- key
String. They key identifies 1) the difficulty estimates pertaining to a particular Rasch model, and 2) the prior mean and standard deviation of the prior distribution for calculating the D-score. The default key
NULLsetskey = "gsed2510". Viewbuiltin_keysfor an overview of the available keys.- population
String. The name of the reference population to calculate DAZ. Use
with(builtin_references, table(key, population))to see which built-in references are available forkey - populationcombinations. If not specified, the function set the default population asbuiltin_keys$base_population[key == builtin_keys$key].- xname
A string with the name of the age variable in
data. The default is"age". Do not round age.- xunit
A string specifying the unit in which age is measured (either
"decimal","days"or"months"). The default"decimal"corresponds to decimal age in years.- prepend
Character vector with column names in
datathat will be prepended to the returned data frame. This is useful for copying columns from data into the result, e.g., for matching.- itembank
A
data.framewith at least three columns namedkey,itemandtau. By default, the function usesdscore::builtin_itembank. If you specify your ownitembank, then you should also provide the relevanttransformandqparguments.- metric
A string, either
"dscore"(default) or"logit", signalling the metric in which ability is estimated.dazis not calculated for the logit scale.- prior_mean
NULL(default), a string, a numeric scalar, or a numeric vector withnrow(data)elements. The default valueNULLwill consult thebase_populationfield inbuiltin_keys, and use the corresponding median of that reference as prior mean for the D-score. The string should refer to a column name indatathat contains user-supplied values of the prior mean for each observation. A numeric scalar will be expanded to all observations. A numeric vector will be used as is.- prior_mean_NA
NULL(default) or a scalar numeric, representing the prior mean for observations with missing ages. By default, D-scores with missing ages will weNA. We suggest settingprior_mean_NA = 50as a reasonable choice for samples between 0-3 years. The argument is ignored ifprior_meanis specified per observation, which gives you full control of priors for observations with missing ages.- prior_sd
NULL(default), a string, a numeric scalar, or a numeric vector withnrow(data)elements. The default (NULL) uses a value of 5 for all ages. The string should refer to a column name indatathat contains user-supplied values of the prior sd for each observation. A numeric scalar will be expanded to all observations. A numeric vector will be used as is.- prior_sd_NA
NULL(default) or a scalar numeric, representing the prior sd for observations with missing ages. By default, D-scores with missing ages will weNA. We suggest settingprior_sd_NA = 20as a reasonable choice for samples between 0-3 years. The argument is ignored ifprior_sdis specified per observation, which gives you full control of priors for observations with missing ages.- transform
Numeric vector, length 2, containing the intercept and slope of the linear transform from the logit scale into the the D-score scale. The default (
NULL) searchesbuiltin_keysfor intercept and slope values.- qp
Numeric vector of equally spaced quadrature points. This vector should span the range of all D-score or logit values. The default (
NULL) createsseq(from, to, by)searching the arguments frombuiltin_keys.- dec
A vector of two integers specifying the number of decimals for rounding the D-score and DAZ, respectively. The default is
dec = c(2L, 3L).- relevance
A numeric vector of length with the lower and upper bounds of the relevance interval. The procedure calculates a dynamic EAP for each item. If the difficulty level (tau) of the next item is outside the relevance interval around EAP, the procedure ignore the score on the item. The default is
c(-Inf, +Inf)does not ignore scores.- algorithm
Computational method, for backward compatibility. Either
"current"(default) or"1.8.7"(deprecated).- verbose
Logical. Print settings.
Value
The dscore() function returns a data.frame with nrow(data) rows.
Optionally, the first block of columns can be copied to the
result by using prepend. The second block consists of the
following columns:
| Name | Label |
a | Decimal age (years) |
n | Number of items with valid (0/1) data |
p | Percentage of passed milestones |
d | D-score, mean of posterior distribution |
sem | Standard error of measurement, standard deviation of the posterior |
daz | D-score corrected for age, calculated in Z-scale (for metric "dscore") |
The D-score in column d is a linear scale, with values usually ranging
from 0 to 100. The D-score is NA if age is missing or if age is lower
than -1/12. It is possible to calculate D-scores for cases with missing ages
by setting prior_mean_NA and prior_sd_NA to some reasonable value, e.g.,
prior_mean_NA = 50 and prior_sd_NA = 20, for the sample at hand.
The SEM is a positive number that quantifies the uncertainty of the D-score.
It is NA if the D-score is NA.
The DAZ in column daz is a Z-score that corrects the D-score for age. It
is NA when there are no reference values for the given age, or when
the D-score is extremely unlikely to be valid at the given age.
Advanced applications: The dscore_posterior() function returns a
data frame with nrow(data) rows and length(qp) plus prepended columns
with the full posterior density of the D-score at each quadrature point.
If no valid responses are found, dscore_posterior() returns the
prior density. Versions prior to 1.8.5 returned a matrix (instead of
a data.frame). Code that depends on the result being a matrix may break
and may need adaptation.
Details
The scoring algorithm is based on the method by Bock and Mislevy (1982). The method uses Bayes rule to update a prior ability into a posterior ability.
The item names should correspond to the "gsed" lexicon.
A key is defined by the set of estimated item difficulties.
| Key | Model | Quadrature | Instruments | Direct/Caregiver | Reference |
"dutch" | 75_0 | -10:80 | 1 | direct | Van Buuren, 2014/2020 |
"gcdg" | 565_18 | -10:100 | 13 | direct | Weber, 2019 |
"gsed1912" | 807_17 | -10:100 | 21 | mixed | GSED Team, 2019 |
"293_0" | 293_0 | -10:100 | 2 | mixed | GSED Team, 2022 |
"gsed2212" | 818_6 | -10:100 | 27 | mixed | GSED Team, 2022 |
"gsed2406" | 818_6 | -10:100 | 27 | mixed | GSED Team, 2024 |
"gsed2510" | 281_0 | -10:125 | 3 | mixed | GSED Team, 2025 |
As a general rule, one should only compare D-scores
that are calculated using the same key and the same
set of quadrature points. For calculating D-scores on new data,
the advice is to use the default, which currently is "gsed2510".
Currently, key "gsed2510" is defined for instrument codes gs1
(GSED SF), gl1 (GSED LF) and gh1 (GSED HF). If you
have another instrument, use the key "gsed2406".
The default starting prior is a mean calculated from a so-called
"Count model" that describes mean D-score as a function of age. The
The Count models are implemented in the function [get_mu()].
By default, the spread of the starting prior
is 5 D-score points around the mean D-score, which corresponds to
approximately 1.5 to 2 times the normal spread of child of a given age. The
starting prior is informative for very short test (say <5 items), but has
little impact on the posterior for larger tests.
References
Bock DD, Mislevy RJ (1982). Adaptive EAP Estimation of Ability in a Microcomputer Environment. Applied Psychological Measurement, 6(4), 431-444.
Van Buuren S (2014). Growth charts of human development. Stat Methods Med Res, 23(4), 346-368. https://doi.org/10.1177/0962280212473300
Weber AM, Rubio-Codina M, Walker SP, van Buuren S, Eekhout I, Grantham-McGregor S, Caridad Araujo M, Chang SM, Fernald LCH, Hamadani JD, Hanlon A, Karam SM, Lozoff B, Ratsifandrihamanana L, Richter L, Black MM (2019). The D-score: a metric for interpreting the early development of infants and toddlers across global settings. BMJ Global Health, BMJ Global Health 4: e001724. https://stefvanbuuren.name/publications/#weber-2019-1
See also
Examples
# using all defaults and properly formatted data
sf <- dscore::triple[, 1:141]
ds <- dscore(sf)
head(ds)
#> a n p d sem daz
#> 1 1.9493 65 0.6769 68.95 1.210561 1.186
#> 2 2.5325 34 0.7059 73.23 1.458141 0.572
#> 3 2.3874 36 0.5833 65.89 1.404537 -0.966
#> 4 0.8980 8 0.5000 38.64 2.527097 -2.228
#> 5 2.1903 31 0.2258 57.84 1.605532 -2.289
#> 6 0.8980 80 0.7625 54.78 1.325298 2.517
# step-by-step example demonstrating
# all possible response vectors for 3 items
data <- data.frame(
id = c(
"Jane", "Martin", "ID-3", "No. 4", "Five", "6",
NA_character_, as.character(8:10)),
age = rep(round(21 / 365.25, 4), 10),
gs1sec001 = c(NA, NA, 0, 0, 0, 1, 0, 1, 1, 1),
gs1moc002 = c(NA, NA, NA, 0, 1, 0, 1, 0, 1, 1),
gs1sec003 = c(NA, 0, 0, 1, 0, 0, 1, 1, 0, 1)
)
# what are these items?
items <- names(data)[3:5]
get_labels(items)
#> gs1sec001
#> "SF001 Does your child smile?"
#> gs1moc002
#> "SF002 When lying on his/her back, does your child move his/her arms and legs?"
#> gs1sec003
#> "SF003 Does your child look at your face when you speak to him/her?"
# difficulty parameter in default key
get_tau(items, verbose = TRUE)
#> key: gsed2510
#> gs1sec001 gs1moc002 gs1sec003
#> 2.56 0.40 3.12
# calculate D-score
# the same sumscore leads to the same D-score (column d)
dscore(data)
#> a n p d sem daz
#> 1 0.0575 0 NA NA NA NA
#> 2 0.0575 1 0.0000 8.22 4.413632 -1.821
#> 3 0.0575 2 0.0000 4.91 3.791544 -2.617
#> 4 0.0575 3 0.3333 5.77 3.547318 -2.424
#> 5 0.0575 3 0.3333 5.77 3.547318 -2.424
#> 6 0.0575 3 0.3333 5.77 3.547318 -2.424
#> 7 0.0575 3 0.6667 9.64 3.953138 -1.426
#> 8 0.0575 3 0.6667 9.64 3.953138 -1.426
#> 9 0.0575 3 0.6667 9.64 3.953138 -1.426
#> 10 0.0575 3 1.0000 14.58 4.470412 0.192
# prepend id variable to output
dscore(data, prepend = "id")
#> id a n p d sem daz
#> 1 Jane 0.0575 0 NA NA NA NA
#> 2 Martin 0.0575 1 0.0000 8.22 4.413632 -1.821
#> 3 ID-3 0.0575 2 0.0000 4.91 3.791544 -2.617
#> 4 No. 4 0.0575 3 0.3333 5.77 3.547318 -2.424
#> 5 Five 0.0575 3 0.3333 5.77 3.547318 -2.424
#> 6 6 0.0575 3 0.3333 5.77 3.547318 -2.424
#> 7 <NA> 0.0575 3 0.6667 9.64 3.953138 -1.426
#> 8 8 0.0575 3 0.6667 9.64 3.953138 -1.426
#> 9 9 0.0575 3 0.6667 9.64 3.953138 -1.426
#> 10 10 0.0575 3 1.0000 14.58 4.470412 0.192
# or prepend all data
# dscore(data, prepend = colnames(data))
# calculate full posterior
p <- dscore_posterior(data)
# check that rows sum to 1
rowSums(p)
#> [1] 0.9999992 1.0000000 1.0000000 1.0000000 1.0000000 1.0000000 1.0000000
#> [8] 1.0000000 1.0000000 1.0000000
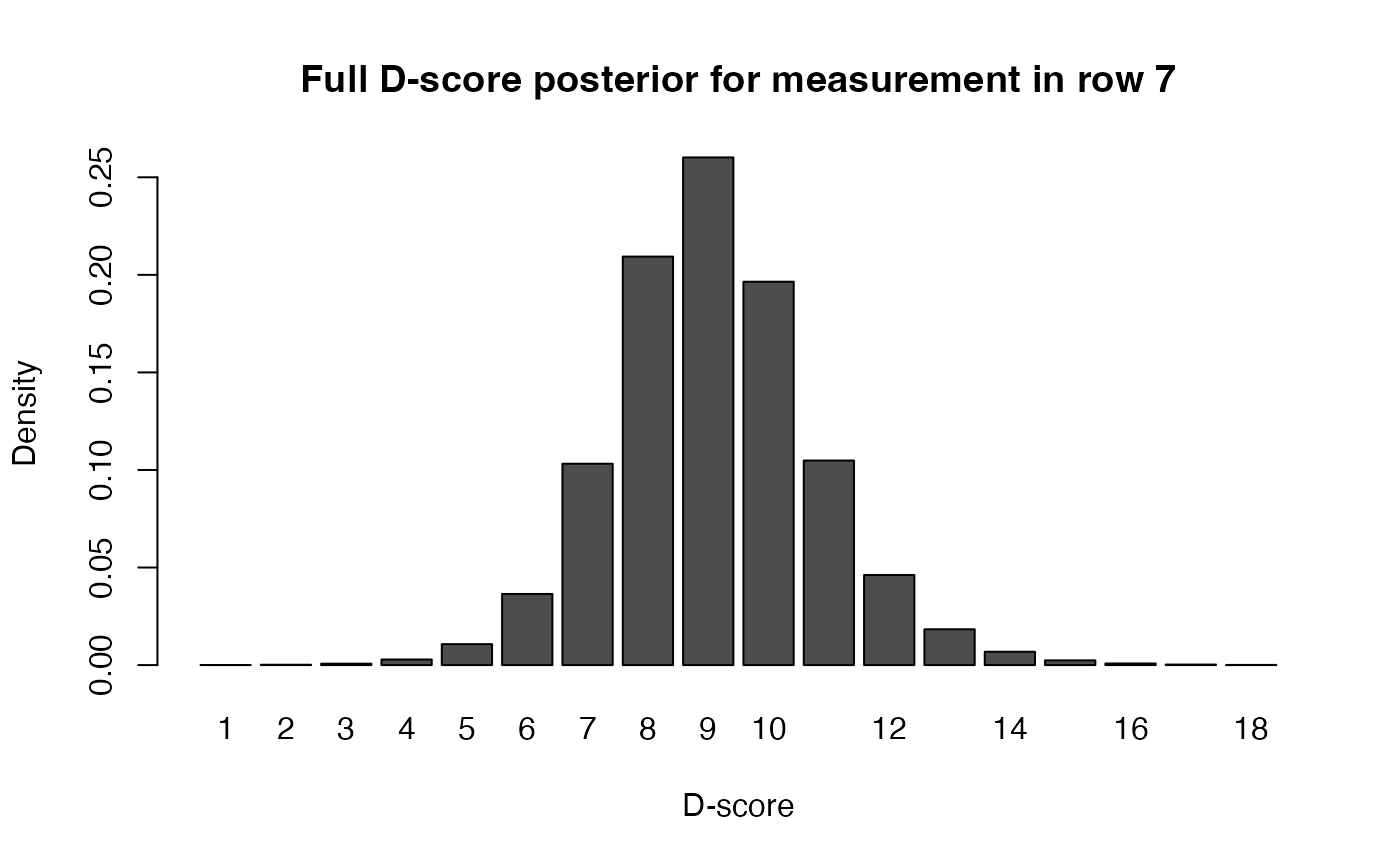
# plot full posterior for measurement 7
barplot(as.matrix(p[7, 12:36]),
names = 1:25,
xlab = "D-score", ylab = "Density", col = "grey",
main = "Full D-score posterior for measurement in row 7",
sub = "D-score (EAP) = 11.58, SEM = 3.99")
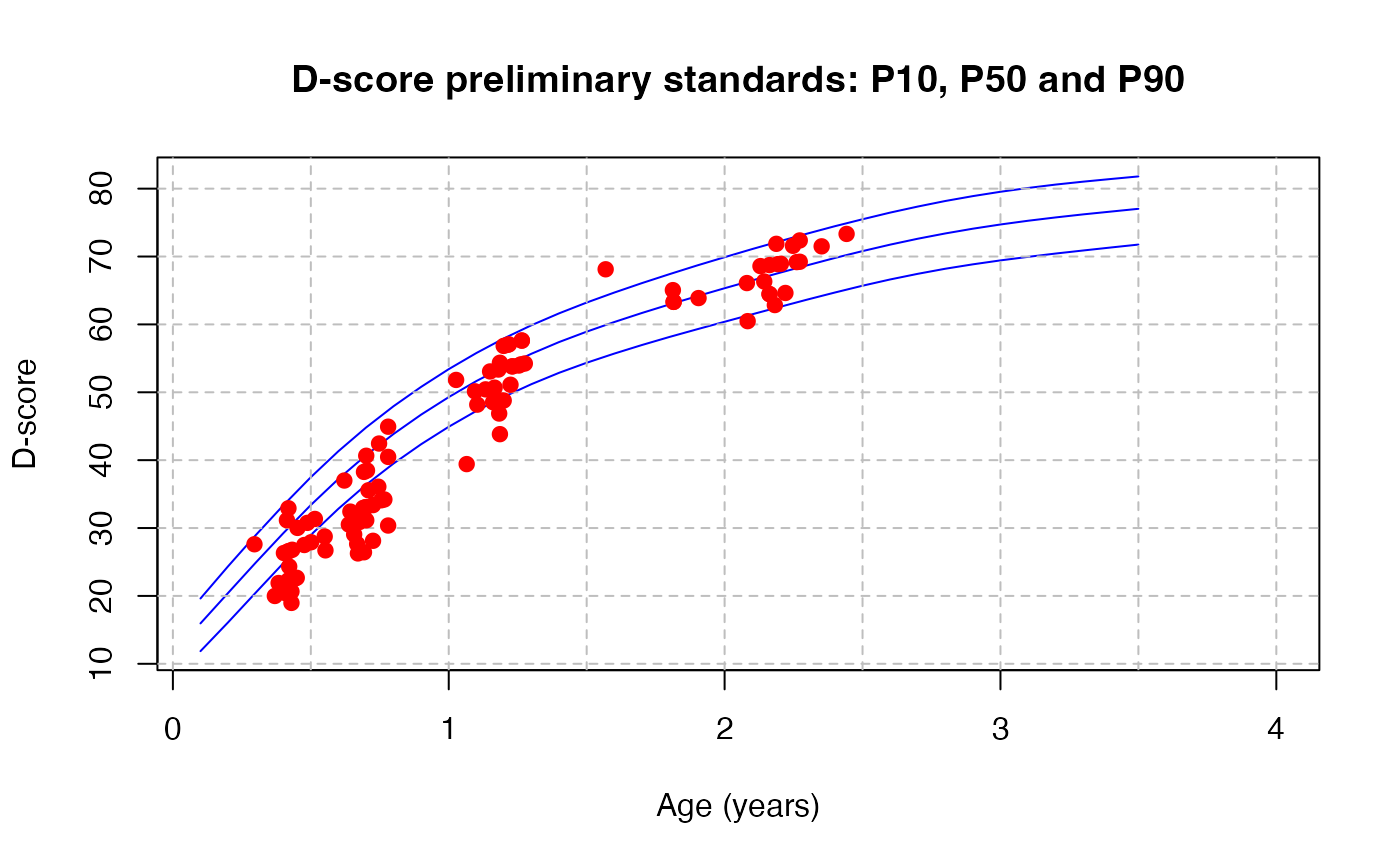
 # plot P10, P50 and P90 of D-score references
g <- expand.grid(age = seq(0.1, 4, 0.1), p = c(0.1, 0.5, 0.9))
d <- zad(z = qnorm(g$p), x = g$age, verbose = TRUE)
#> key: gsed2510
#> population: preliminary_standards
matplot(
x = matrix(g$age, ncol = 3), y = matrix(d, ncol = 3), type = "l",
lty = 1, col = "blue", xlab = "Age (years)", ylab = "D-score",
main = "D-score preliminary standards: P10, P50 and P90")
abline(h = seq(10, 80, 10), v = seq(0, 4, 0.5), col = "gray", lty = 2)
# add measurements made on very preterms, ga < 32 weeks
# we need key = "gsed2406" because DDI is not yet in key "gsed2510"
ds <- dscore(milestones, key = "gsed2406")
points(x = ds$a, y = ds$d, pch = 19, col = "red")
# plot P10, P50 and P90 of D-score references
g <- expand.grid(age = seq(0.1, 4, 0.1), p = c(0.1, 0.5, 0.9))
d <- zad(z = qnorm(g$p), x = g$age, verbose = TRUE)
#> key: gsed2510
#> population: preliminary_standards
matplot(
x = matrix(g$age, ncol = 3), y = matrix(d, ncol = 3), type = "l",
lty = 1, col = "blue", xlab = "Age (years)", ylab = "D-score",
main = "D-score preliminary standards: P10, P50 and P90")
abline(h = seq(10, 80, 10), v = seq(0, 4, 0.5), col = "gray", lty = 2)
# add measurements made on very preterms, ga < 32 weeks
# we need key = "gsed2406" because DDI is not yet in key "gsed2510"
ds <- dscore(milestones, key = "gsed2406")
points(x = ds$a, y = ds$d, pch = 19, col = "red")